1 Getting started with R
(MA7419 / MA3419)
1.1 Overview
Objectives
On successful completion of this module, you should be able to:
- Define data science and discuss its role in actuarial science and business analytics
- Create and run simple program scripts in the RStudio environment and read data into R from a local tabular file
- Create and share literate programs and reports using R Markdown and undertake simple EDA (including summary statistics and informative visualisations) of data in a tabular structure
- Implement a reproducible workflow for simple data science project and describe ethical and regulatory issues to be considered when undertaking a data science project
I prefer an alternative statement of the objectives:
By the end of this module you will be confident in handling data in a collaborative and reproducible way. In other words in a professional way. You will be able to:
- get
- clean
- check
- restructure
- filter, sort, join & summarise
- explore
- visualise…
…data
The specific objectives for this week are:
- Access your R Server account
- Upload Task 1 from Blackboard to the server
- Complete the exercises in the Week 1 task
- Knit to HTML and download HTML and RMD files
- Complete the Week 1 Blackboard test
- Submit RMD and HTML files on Blackboard
- Complete the Week 1 Reflective Survey
Problem-based learning
The structure of this module may be a bit different to what you have come across before.
In a “traditional” university lecture you arrive without having done any preparation, the lecturer gives you some information, and you go away to try and absorb it and probably do some exercises or homework.
In a “flipped” classroom you are supposed to have done reading, or used other resources, so that you have some familiarity with the material before the class. In class you focus on applying what you have learnt to some exercises.
This module is quite similar to a flipped classroom, but here, working on the task and learning what you need to successfully complete it will happen much more in parallel. I will give you a quick overview of some of the things you need to know each week, and there will be some indicative reading. But after that it is up to you (usually working as part of a team) to work out a good solution to the task.
This is much more like the way things happen in real life.
Introductions
- Dr Paul King FIA (Lecturer)
- paul.king@leicester.ac.uk
- No set office hours: email for an appointment
- Your friendly TAs
- Silvia Freund - skf10.le.ac.uk
- Ibrahim Issahaku - iai5@le.ac.uk
Class times
- Tuesday 12:00 - 13:00, Rattray Lecture Theatre (please enter through upper doors)
- Wednesday 11:00 - 13:00, Computer Labs
- Charles Wilson 304/305 for undergraduates and ASDA
- Sir Bob Burgess 0.04 for DABI
How this module works
- A certain amount of lecturing (mostly on Tuesdays)
- A lot of group problem solving (on Wednesday)
- A lot of self study: using the reading list and other on-line resources
- Additional resources:
- Data Science Club (if you want to run one!)
- DataCamp
- The weekly tasks for an important part of the learning activity on this module and the completed tasks will be a key part of the set of notes you develop
Group working
From next week you will be working in groups on a weekly coding task. The output will be a R Markdown notebook containing your code and results - each member of the group must produce their own notebook and be able to edit it and run it on the server or their own machine. Completed tasks (i.e. the PDF file) should be submitted by midnight on Sunday (UK time) in the week of the class.
Getting help on the weekly task
You can ask for help from your TA during the Wednesday computer lab. Outside of the lab (or even inside) you can post questions on the Blackboard discussion forum. Try to formulate your question as specifically as possible and give a code sample. That way the question can be answered in the thread and benefit everybody (other students are encouraged to answer questions). If you need more detailed help a TA will contact you via Teams.
I’ll create a separate chat for general questions about the course. Please try to put as many of your questions here as possible so everyone can benefit, but if it’s something a bit more personal, please feel free to email me (paul.king@leicester.ac.uk) directly. I’m happy to arrange a one to one meeting over Teams or face to face.
Submitting your weekly work
When you’ve completed the weekly task, please submit both your RMD and HTML (sometimes you will be asked for PDF) files via the assignment links on Blackboard.
You should also complete the short weekly Blackboard test. This is based on the answers in the exercises, but will sometimes require some extra thinking or research.
Finally, you should complete the weekly reflective survey. This is designed to act as a prompt for you to reflect on your week’s work. It also gives us a useful indicator of who might need extra help.
Links to the test and the survey are in the Blackboard section (the Blackboard term is a Learning Module).
All of the above are important “formative” activities to help you learn, give you feedback on your progress, and motivate you to keep up with the work. The School will use them as part of the way it monitors your engagement with the module.
However, none of the marks count toward the final assessment for the module. (Assessments that count for that are called “summative”.)
DataCamp
There’s a link to register in the Blackboard site, and there is more information in Getting Set Up.
It’s a very good idea to take advantage of this resource while it’s available.
How this module is assessed
- Coursework mid-semester (30%)
- similar to problems solved in class; but
- you must work on your own
- available at the end of week 15 and due in two weeks later
- Final report and presentation (70%)
- you will work on a group data science project and produce both a detailed report of the analysis and a presentation
- the presentation must include slides (generated from R markdown) but they will be marked on the submitted document - you will not be required to give a stand up presentation
- each individual will have to identify an aspect they worked on, for questions
- some of the marks will be for demonstrating knowledge of the topics in the lectures after revision week
- assessments for MA3419 and MA7419 are different
1.2 Reading
R for Data Science (Wickham and Grolemund 2017):
- Chapter 4 Workflow: basics
R Programming for Data Science (Roger D. Peng 2020):
- Chapter 13 R nuts & bolts
Your own research
1.3 Reproducible data science
Introduction
We are going to discuss Reproducible Data Science. This is often referred to as Reproducible Research, but it applies to lots of data science activities we might not consider as research.
It’s helpful to consider the process of data science, for example as illustrated by Roger Peng in his book Report Writing for Data Science in R (Roger D. Peng 2019).
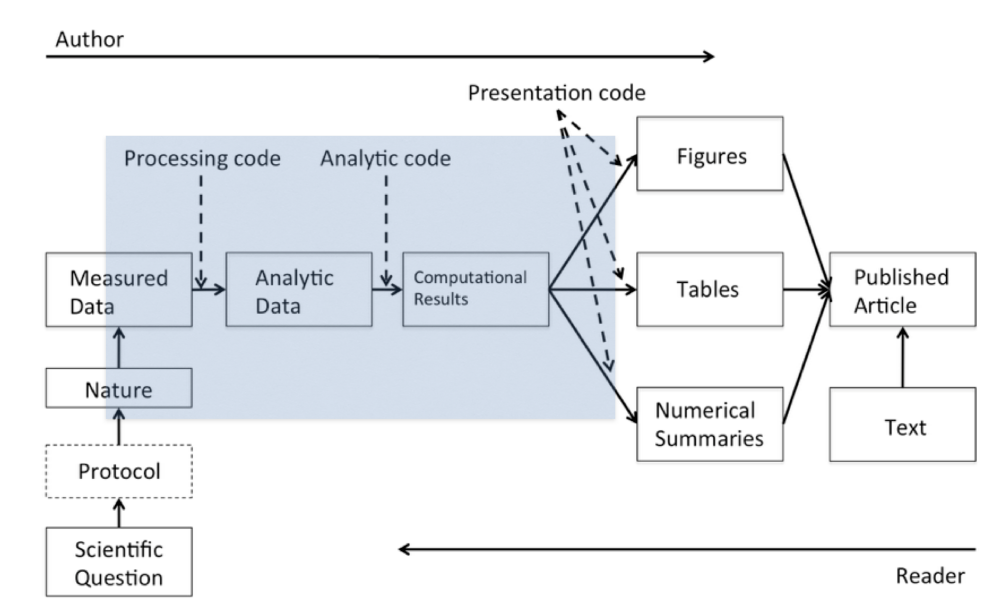
The steps in the shaded box start with the raw (measured) data and end with the computational results. In this module we’ll be concerned with these steps - the core of a reproducible pipeline - plus the presentation code used to produce the final output.
In this diagram analytic data refers to the raw data after it has been checked, cleaned and reshaped into the right format for further analysis.
What is Reproducible Data Science?
A reproducible data science process allows a reviewer or subsequent user of the work to reproduce the analysis at least from the analytic data to the computational results. It should also allow them to:
- see what has been done at each stage;
- understand the reasons for the decisions made at each step;
- check that the claimed steps have been carried out correctly.
In all but the simplest of projects, this requires the publication of well written, and well documented, code.
When we consider published work in the context of a scientific publication, reproducibility, as a minimum, requires making available the analytic data, code and details of the computational environment used.
However, it’s important the the whole process, including pre-processing the data and producing the final outputs, is fully documented.
Benefits (and disadvantages) of Reproducible Data Science
Scientific:
- Replication of published work is a key element of the scientific process: reproduction of the analysis of data analysis is often the closest to replication it’s realistically possible;
- attempts at reproduction can reveal errors with potentially serious consequences.
Professional:
- Ability to reproduce a workflow is key to checking for errors;
- Documentation of data analysis is required for work review and auditability;
- For actuaries, the professional standards APS X2 and APS QA2 require consideration to be given to reproducibility in relation to the above points;
- A reproducible work flow allows efficient reuse or modification of an analysis pipeline.
Personal:
- The next user of your work is most likely to be you!
- the process of making your work reproducible requires you to stop (or at least slow down) occasionally to make sure you are working effectively and efficiently.
How to make sure your work is reproducible
- Create a consistent filing structure for data, code, intermediate and final results;
- see the next section
- Use RStudio and
knitrto practice literate programming by creating R Markdown notebooks; - Resist the temptation to do things manually - do as much programmatically as possible
- coding all your data manipulation is not just about automating it, it’s about documenting what you’ve done so it can be reproduced;
- Comment code appropriately;
- Stick to a coding standard (style guide)
- we will use the tidyverse style guide
- Save as few intermediate results as possible;
- Use version control;
- Keep track of your programming environment
The last two points are important, but we don’t have the time to go into them in any detail in this course.
Further Reading
APS X2 Review of Actuarial Work
APS QA1 Quality Assurance for Organisations (Version 2.0)
The Institute and Faculty of Actuaries has written about how important Data Science is to the profession.
1.4 More resources
The Bookdown home page gives you access to many very good books covering different aspects of data science with R.
1.5 Setting up your R environment.
You will be working in R every week during this module, so the first task is to make sure you have access to RStudio.
The next step is to configure your environment in a standard way so it will be relatively straightforward for us all to work share code and work together.
Go ahead and:
- Use RStudio to create a project called MA3419 or MA7419
- Inside that project, create folders for each Week during the Semester, plus a folder called Data
- here is a video demonstrating how to do steps 1 & 2.
- Install the packages ‘tidyverse’ and ‘here’
- video demonstration
- Get familiar with the RStudio interface
- Get familiar with the R Markdown notebook structure and how to run code inside a notebook
- video
- I highly recommend this blog article
- if you need more help, you’ll almost certainly find something at RStudio
- once you’re reasonably familiar with RStudio and R Markdown have a look at some more advanced Tips and Tricks (This won’t make much sense if you’re an absolute beginner, but you should bookmark it and come back to it later.)
1.6 Week 1 Task
Every week there will be a task provided as a .RMD file in the Blackboard folder. You need to copy this file into the appropriate week’s folder in your MA3419/MA7419 folder and have it open in a running R session during the Wednesday computer lab. The task files will become available on Monday mornings.
The marks for the weekly task are for feedback only. They don’t count towards your module assessment.
Sometimes there will also be a data file supplied - you should copy this into your project’s Data folder.
Sometimes I’ll work through parts of the task during the Tuesday lecture, and every week I’ll give feedback on the most common mistakes in the previous week’s task. There will also be exercises embedded in the notes for you to complete.
Your target should be to complete all the exercises in each weekly task, and submit your answers on Blackboard. You should also do the week’s Blackboard test for feedback
I’ll release the correct code for the previous week’s task on Monday morning, this will include feedback notes based on the submissions where I think that will help.
Occasionally there will be challenge exercises or challenge tasks - these are optional.
1.7 Addendum 1: R Markdown vs Quarto
As you reseach material online you’ll come across references to both R Markdown and Quarto. You’ll also see the option to create Quarto documents (along with a lot of other options) when you start a new file in R Studio.
R markdown is the original format which is gradually being replaced by Quarto.
For our purposes there is very little difference and we’ll stick with R Markdown.
You can try it for yourself. Go to File > New File and create two new files: one using R Markdown, and one using Quarto Document.
Look at the starter code provided (you don’t need to save them) and try to spot the difference.
You are welcome to use the Quarto format when you create your own new files, but the weekly tasks are in R Markdown format and we’ll stick with that for now. (Although these notes are actually written in Quarto.)
1.8 Addendum 2: R vs Python
Python is an important language for data science and machine learning and I always get asked if you can use Python instead of R.
The short answer is “no”, because one of the objectives of this module is for you to learn R.
However, I’m keen to explore the possibility of creating a Python version of this course so if you are interested on working on creating Python code to reproduce the examples and tasks (perhaps as part of a Data Science Club), please email me.